ISHpalette™ Short hairpin amplifier is a fluorescent hairpin DNA reagent developed as an enhanced version of the traditional in situ HCR method for detecting target molecules in tissue sections.
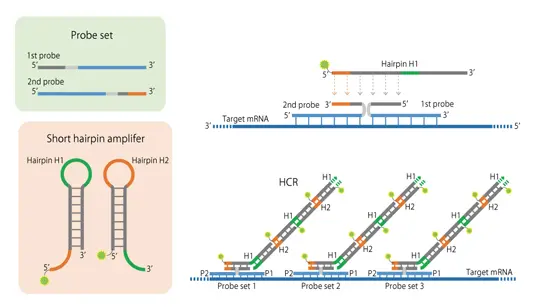
The in situ Hybridization Chain Reaction (HCR) method utilizes the self-catalyzed elongation of nucleic acids with a hairpin structure 1) to initiate a chain polymerization reaction of labeled DNAs on tissue sections, thereby enabling visualization of target nucleic acids 2).
Because this method does not rely on antibody or enzymatic reactions, it exhibits linear signal intensity and a high signal-to-noise ratio.
The hybridization of two probes with sequences that initiate the HCR (polymerization reaction) of the hairpin DNAs, followed by the subsequent HCR, results in fluorescently labeled DNAs specifically binding to the target mRNA.
HCR occurs only when the two probes hybridize in the correct positions, minimizing nonspecific reactions. The ISHpalette™ is a further shortened version of the hairpin DNA, eliminating the need for the conventional permeation treatment 3). Furthermore, because the hybridization time per molecule is reduced, the reaction reaches plateau faster, resulting in higher signal intensity.

The mRNA of Vglut2 (green), Sik3 (red), and Vgat (magenta) were detected on mouse brain sections. Each mRNA molecule can be detected at a single granule.

The c-Fos protein was detected by immunostaining (left, green), and Vglut2 mRNA was detected by in situ HCR (center, magenta) on mouse brain sections. In the Proteinase K-treated (+) samples (top row), faint c-Fos protein signal was detected, whereas in the untreated (−) samples (bottom row), both the protein and mRNA were clearly detected simultaneously. (Modified from Tsuneoka and Funato, 2020, Front. Mol. Neurosci.)
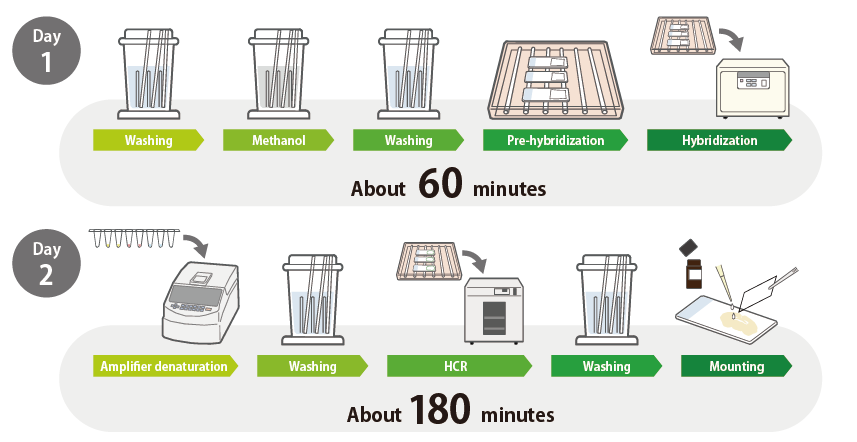
Flowchart: in situ HCR with ISHpalette™ for Frozen Sections
No special equipment or reagents required.
Permeabilization treatments with Proteinase K, acetylation, high-temperature treatment, or RNase treatment—are not necessary, resulting in a simplified protocol and faster staining process.
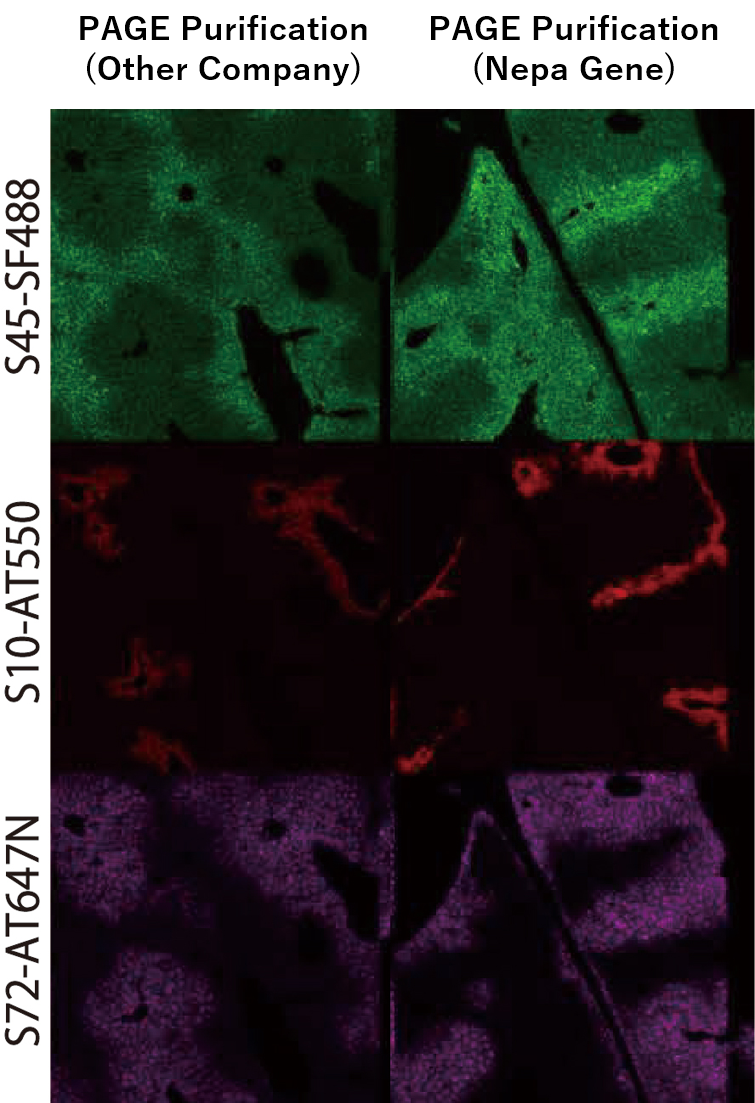
A comparison of staining performance was conducted between fluorescently labeled hairpin DNA purified by a general oligo manufacturer (standard PAGE purification) and our proprietary PAGE-purified product. Our original product, developed in collaboration with Toho University, demonstrated higher fluorescent labeling efficiency and improved reactivity, resulting in stronger fluorescence signals. The comparison was performed using mouse liver tissue targeting Albumin, Cyp2e1, Glul.
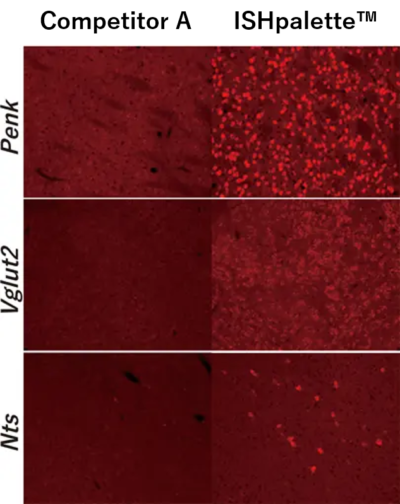
The mRNA of Penk (top), Vglut2 (middle), and Nts (bottom) in mouse brain sections was detected using both the conventional HCR method and ISHpalette™. Signal intensity was compared between the two approaches. The results clearly show that ISHpalette™—our shortened hairpin DNA—provides improved tissue permeation and reactivity. All were stained without Proteinase K (ProK) treatment.
| Item No. | Description |
| IPL-G-S23 | ISHpalette™ Short hairpin amplifier, SaraFluor™488-S23 (25 slides: 50μL each) |
| IPL-G-S45 | ISHpalette™ Short hairpin amplifier, SaraFluor™488-S45 (25 slides: 50μL each) |
| IPL-R-S41 | ISHpalette™ Short hairpin amplifier, ATTO550-S41 (25 slides: 50μL each) |
| IPL-R-S73 | ISHpalette™ Short hairpin amplifier, ATTO550-S73 (25 slides: 50μL each) |
| IPL-B-C-S72 | ISHpalette™ Short hairpin amplifier, Cyanine5-S72 (25 slides: 50μL each) |
| IPL-B-C-A161 | ISHpalette™ Short hairpin amplifier, Cyanine5-A161 (25 slides: 50μL each) |
→Here is the sequence information for each hairpin DNA (Excel: 11KB).
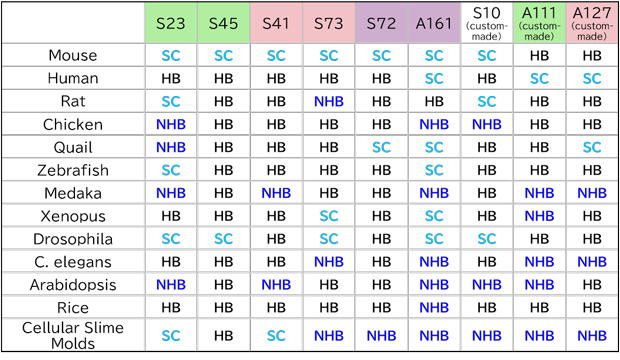
We accept custom orders for combinations of hairpin DNAs and labels that differ from our standard products, custom-made hairpin DNAs (refer to the species compatibility table below), and other labeled products (such as biotin, ATTO647N, etc.).
| Item No. | Description |
| IPL-HB | ISHpalette™ Hybridization buffer (30mL) |
| IPL-AB | ISHpalette™ Amplification buffer (30mL) |
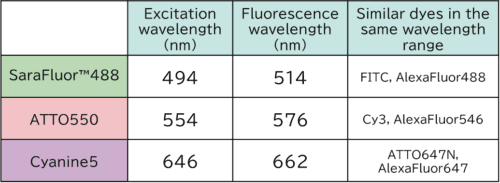
SaraFluor™ 488: A dye manufactured by a Japanese manufacturer, known for its resistance to photobleaching. Here is the page for SaraFluor™ 488.
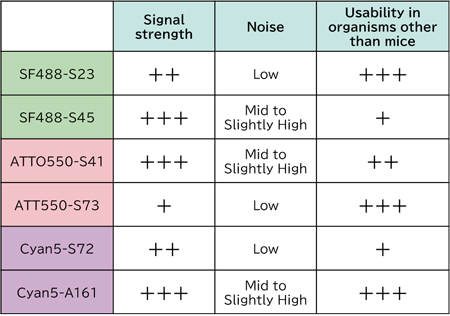
| SC (Staining confirmed) | Staining has been successfully performed to date, with little to no non-specific reactions observed in the tested tissues. |
| NHB (No Hits found in BLAST) | No mRNA sequences predicted to cross-react with the hairpin initiator were found via BLAST search. Non-specific reactions are considered unlikely. |
| HB (Hits found in BLAST) | Potential mRNA sequences that may cross-react with the hairpin initiator were identified in a BLAST search. Non-specific reactions could occur depending on developmental stage or tissue type. |
Note: Studies on other species are currently ongoing. You can proceed with using the product after confirming through negative controls that there are no non-specific reactions.
For custom-made hairpin DNA (S10, A111, A127), please contact us.
→Here is the assessment protocol for evaluating hairpin DNA usability.
We offer custom probe design services. Based on the target sequence, we select the optimal probe sequences designed with consideration of factors such as Tm values and GC content. The final deliverable includes the probe sequence information along with the binding sequence for the hairpin DNA.
| Item No. | Description |
| IPL-DSGN-PB | Custom design of target probe (1 target). |
Note:
Using the provided sequence information, you will need to order the oligonucleotides from a synthesis company on your own.
If you wish to design the probe yourself, please refer to the user manual.
→An example of a designed probe (positive control: Gapdh) is available here (Excel: 10KB).
For each region, two types of target probes are designed, one for the 5′ side and one for the 3′ side. Detection cannot be achieved with only one type of hybridization, so you can use a probe with only one of the target probes for the 5′ or 3′ end as a negative control.
Note:

TiYO™ Autofluorescence Quenching Illuminator
訊詢單內容